Introduction to the -omics (Genomics, Transcriptomics, Proteomics, Metabolomics)
Biological systems can be studied at a number of levels, starting from DNA and working upwards towards life-sustaining reactions. The most fundamental level would be genomics, the study of DNA structure, function, modification, and utilization. This area gives us insight into the instructions that code the lives of ourselves and the natural world around us. Transcriptomics, the study of RNA concentrations within a system, gives us an idea about what cellular activities are prioritized or suppressed within a living system. Proteomics, the study of proteins, can tell us the composition and location of proteins within a living system. The analysis of each of these levels has certain limitations. For example, the study of DNA tells us only what could happen, as the presence of a sequence in the DNA does not mean that it will be expressed. The study of RNA tells us about what is intended to happen, as these would code for the proteins that the cell wants to produce; however, more than half of all proteins undergo post-translational modifications, and these are not accounted for in the mRNA that codes the proteins primary sequence. Proteins activity is also not directly correlated to transcription rates, making mRNA abundance an unreliable detector of said activity. Finally, just because a protein is present in the organism, does not mean it is active; as previously stated, many proteins undergo post-translational modifications, and these can be utilized to activate or deactivate these proteins. Additionally, some proteins form complexes with other proteins or RNA molecules, and it can be difficult to account for these interactions in a system-wide study of protein concentrations.
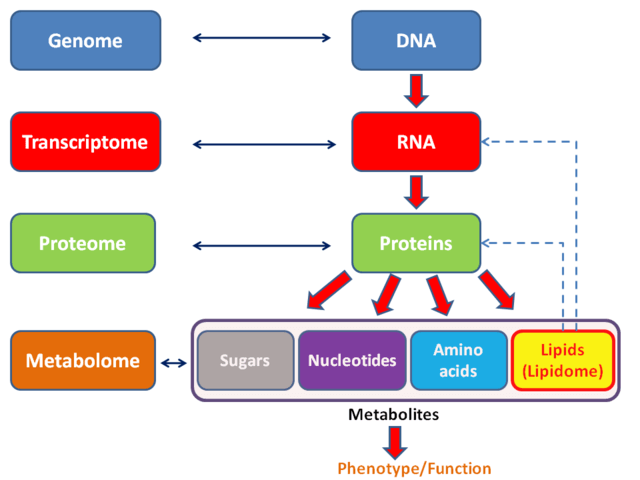
The final level of study would be the analysis of small molecules within a system, and the change in their presence and concentration over time; thus, it is the study of chemical processes within a biological system. This field of study is called metabolomics, and it is unique in that one would not be looking at the actual biological machinery to infer and rationalize activity, but at the small molecules that make up the starting materials, intermediates, and products of these biological processes. A comprehensive analysis of all small molecules present within a system, both at one time and over time, can give incredible insights into cellular, tissue, and organism health.
Metabolomics
Metabolomics is the study of all chemical processes involving metabolites that are occurring within a biological system. Metabolites are small molecules that make up the substrates, intermediates, and products of cellular processes. The study of metabolites and the metabolome, a complete set of all small molecules found within an organism, is exceedingly challenging, as a gargantuan variety of metabolites are present at a wide range of concentrations. Additionally, the cellular matrices for metabolomics analysis varies widely; examples of common samples utilized for metabolomics studies are cell and tissue digests, blood serum and plasma, urine, saliva, feces, sweat, exhaled breath, as well as gastrointestinal fluid. Each of these may require a unique extraction and purification procedure to isolate the desired set of metabolites.
Metabolome
The metabolome refers to the complete set of small molecules found within a specific biological system, from an organelle all the way to a complete organism. A small molecule is defined as being <1.5 KDa (<1,500 g/mol) in molecular mass. Exceptions to this rule exist, as some lipids and albumin are counted towards the metabolome. As the metabolome is constantly changing and consists of a large variety of molecules, both in size, structure, and functionality, no single analytical technique and sample processing procedure accounts for all small molecules within a system.
A number of efforts exist to categorize the metabolome, both in humans and other species. METLIN, created by the Siuzdak laboratory at the Scripps Research Institute in 2003 and made publicly available in 2005, is the largest repository of tandem mass spectrometry data gathered experimentally from genuine standards. There are over 450,000 metabolites or other chemical entities that have experimentally determined mass spectrometry data, and this service has over 30,000 registered users. These metabolites and small molecules encompass a wide range of biological systems, not just human. For humans specifically, the first draft of the human metabolome was published in 2007 as a joint-research effort between the University of Calgary and the University of Alberta, both situated in the province of Alberta, Canada. 2500 metabolites, 1200 drugs, and 3500 food components found within the human body were characterized; each one of these components has, on average, 90 different entries, encompassing NMR and MS spectra, structural information, biofluid concentrations, disease associations, etc. This work can be found under the Human Metabolome Database, and is constantly being updated.
Metabolites
As mentioned previously, metabolites are defined as small molecules found within biological system with a molecular mass <1.5 KDa. Some exceptions exist to this rule, as some lipids and albumin count as metabolites, though they are >1.5 KDa in mass. Metabolites can be split into different categories.
Primary metabolites are small molecules that are directly concerned with the normal growth, development, and reproduction of an organism. Examples of these would include proteins, lipids, carbohydrates, and vitamins. Secondary metabolites are indirectly associated with growth and development, but have important ecological and fitness purposes, as they can stimulate growth or protect the organism from harm. A human example of a secondary metabolite would be hormones. Plant secondary metabolites, used in a defensive or signaling capacity, have found a variety of uses by humans as medicines, flavoring agents, pigments, and recreational drugs.
Metabolites can also be classified as endogenous or exogenous. This classification set is more commonly used for human-originating metabolites. Endogenous metabolites are produced by the organism itself; examples of these would be any of the intermediates found within the citric acid cycle in aerobic cellular respiration. Exogenous metabolites would be small molecules not produced by or expected to be found within the organism, so metabolites of outside origin. These can include drugs, environmental contaminants, toxins, and food additives. Human-produced metabolites that can be traced back to exogenous metabolites (for example, by-products of drug metabolism) are termed xenometabolites.
Automation in Metabolomics
Analytical methods, including separation and detection protocols, for metabolites will be discussed in a future blog-post. However, it can still be said that the techniques required for extracting, purifying, and concentrating metabolites from complex biological samples can be a labor-intensive and time-consuming task. Higher throughput metabolomic centers, including academic core facilities, are turning towards increasing automation to streamline sample processing. As mass spectrometry is a major technique used in the identification of metabolites, we provide a number of solutions in the area of automated mass spectrometry sample preparation, including automated solid phase extraction system and liquid-liquid extraction workstations.


