We all have to get an annual flu shot. I know, it’s not really fun, but at least you’ll be protected during flu season, and hey, if your nurse is nice maybe you will get a lollipop afterwards! But why do we have to get a flu shot every year? Most of us know that this is due to the changing viral coat proteins, specifically hemagglutinin and neuraminidase (the H and N when you talk about flu strains), which makes it so our antibodies no longer recognize the virus and no longer protect us from infection. Thus, like it or not, we have to deal with our upper arm being sore at least one day a year. But why does this happen?
In general, viruses undergo a higher mutation rate than bacteria or other pathogens. For flu, this is in part due to high viral loads. Viral particles can mutate at up to ~2×10-6 changes per generation at viral loads of up to 1010 copies per mL; this would give approximately 100,000,000 mutants for every replication cycle. While our immune system can eliminate most of these “new” viral particles, some are passed to new hosts and can become new strains in future years.
This system of changing your surface proteins (otherwise known as antigenic variation) is by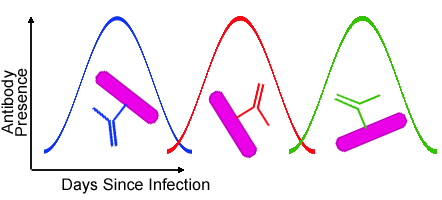 no means unique to viruses. The bacteria which cause meningitis and gonorrhea (Neisseria spp.) are capable of modifying the sequence of their pilin gene, a major immune target in mammalian infection. Additionally, Borrelia burgdorferi, the bacteria which causes Lyme disease, can actively “mutate” the surface protein VlsE by recombining sequence from “silent” loci into a single expressed locus to change the structure of the vlsE protein and escape immune recognition. Using this antigenic variation strategy, B. burgdorferi is able to evade the immune system and remain in the host indefinitely; the bacteria constantly stay ahead of the immune system, as one population of the bacteria is killed off, smaller variant populations which are not yet targeted by the immune system undergo clonal amplification. Talk about natural selection in action!
no means unique to viruses. The bacteria which cause meningitis and gonorrhea (Neisseria spp.) are capable of modifying the sequence of their pilin gene, a major immune target in mammalian infection. Additionally, Borrelia burgdorferi, the bacteria which causes Lyme disease, can actively “mutate” the surface protein VlsE by recombining sequence from “silent” loci into a single expressed locus to change the structure of the vlsE protein and escape immune recognition. Using this antigenic variation strategy, B. burgdorferi is able to evade the immune system and remain in the host indefinitely; the bacteria constantly stay ahead of the immune system, as one population of the bacteria is killed off, smaller variant populations which are not yet targeted by the immune system undergo clonal amplification. Talk about natural selection in action!
Eukaryotic parasites can also utilize antigenic variation strategies to evade the host immune system. The malaria parasite, Plasmodium falciparum, is able to vary one of its surface proteins, PfEMP1, so that infected cells are not recognized by the host immune system. Trypanosoma brucei, which causes African sleeping sickness, can change its variant surface glycoprotein (VSG) through a combination of differential expression and recombination of silent genes into an expressed locus.
Recently, next generation sequencing (NGS) has been applied to help understand the dynamics of antigenic variation in Trypanosoma brucei. During infection in mice, NGS library preparation was carried out by taking blood samples and amplifying expressed vsg RNA using primers specific for the conserved 5’ and 3’ sequences. NGS analysis revealed that multiple different vsg alleles were expressed at any given time in the population, however, allelic frequency decreased at specific time points, evidently as antibodies were produced to target this new clonal population.
Our understanding of antigenic shift and antigenic variation is key to producing effective vaccines; the flu vaccine must be administered every year while no effective vaccines exist for Lyme disease or malaria. New technologies, such as next generation sequencing, are making it possible to examine large, complex populations with relatively routine experiments to aid our understanding of how these variants arise, and could potentially aid in prediction of new variant sequences. Imagine only going for a flu shot once every ten years, or being immunized once in your lifetime. Our ability to reach these goals is rapidly improving; new technologies are pushing our understanding of life, how it works, and our ability to treat diseases. Next generation sequencing has revolutionized our ability to perform research, and is poised to revolutionize diagnosis and patient care. What is the next technology that will change the world?


